Spring papers news
 We enjoyed a wonderful spring flower season here in Southern California
We enjoyed a wonderful spring flower season here in Southern California
(Harford Springs Riverside County Regional Park)
Several publications were accepted and published in last few months and want to highlight the hard work from lab members and collaborators.
Jinfeng lead article published on the localized burst of Rice Transposable Elements Ping and Pong in only a subset of the domesticated lineages and clues to what changed to drive their increased activity.
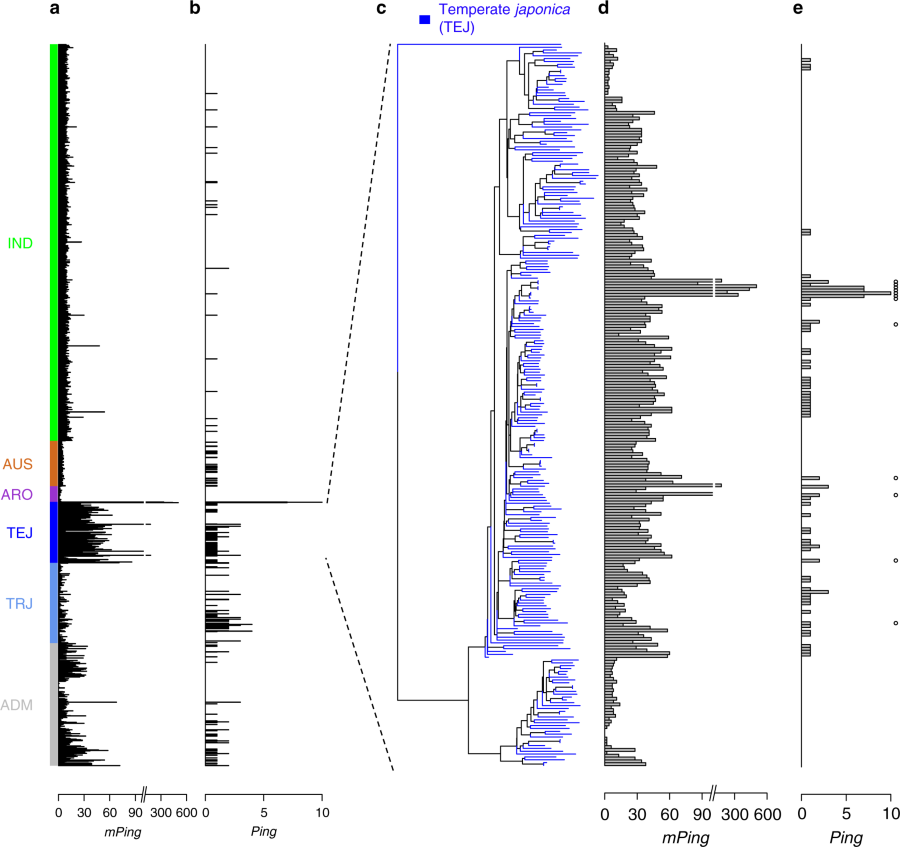 Variation of mPing, Ping, and Pong_ element copy number in rice subgroups https://www.nature.com/articles/s41467-019-08451-3/figures/2
Variation of mPing, Ping, and Pong_ element copy number in rice subgroups https://www.nature.com/articles/s41467-019-08451-3/figures/2
A fun collaboration lead by Adam Bewick and Bob Schmitz we called #MycoMeth explored diversity of Cytosine DNA Methylation in Fungi looking at the 5-methylcytosine (5mC) marks and evolution of the fungal 5mC methyltransferases.
 Genomewide 5mC profiles https://www.nature.com/articles/s41559-019-0810-9/figures/2
Genomewide 5mC profiles https://www.nature.com/articles/s41559-019-0810-9/figures/2
Two Zygolife project papers. One on genome evolution and phylogenic position of Endogone - a truffle forming Mucoromycotina fungus.This was quite challenging but interesting project to tackle, lead by Ying Chang in the Spatafora Lab. Dr Chang took a metagenomics approach to capture these unculturable fungi followed by bioinformatic analyses to isolate the fungal and Mucoromycotina component of the dataset.
 <figcaption>Phylogenetic placement and divergence time estimation of Endogonales and the evolution of fungal plant‐cell‐wall‐degrading enzymes (PCWDEs)
<figcaption>Phylogenetic placement and divergence time estimation of Endogonales and the evolution of fungal plant‐cell‐wall‐degrading enzymes (PCWDEs)
A second paper lead by William Davis from Tim James lab on the genomes and phylogeny of Zoopagomycotina fungi which include nematophagous and mycoparasites.
 Fig 5. Ancestral state reconstructions of trophic state on the best ML tree (A) and the ASTRAL consensus tree (B) using maximum likelihood. Pie graphs at nodes show the proportional likelihood of each character state.
Fig 5. Ancestral state reconstructions of trophic state on the best ML tree (A) and the ASTRAL consensus tree (B) using maximum likelihood. Pie graphs at nodes show the proportional likelihood of each character state.
Jason participated in a Marine Fungi workshop on the unexplored diversity in the marine environments and co-authored this perspective on the opportunities and challenges in characterizing and studying these fungi.
 Morphological diversity of fungi collected from a biotic host. Fungal collection isolated from a marine sponge,Ircinia variabilis (formerly Psammocinia sp.).
Morphological diversity of fungi collected from a biotic host. Fungal collection isolated from a marine sponge,Ircinia variabilis (formerly Psammocinia sp.).
Former postdoc Ousmane Cissé developed FGMP, a tool for assessing completeness of fungal genomes which uses both conserved protein coding markers that can include multicopy gene families and conserved non-coding DNA regions. This was published this month as well, congrats Ousmane!
 Fig 2.Estimation of genome completeness in fungal genomes. A comparison of genome completeness by multiple software tools using initial and latest genome assembly versions of 45 fungi. Genome completeness expressed as a percentage of expected markers (y-axis) is plotted against assembly size in megabases (x-axis).
Fig 2.Estimation of genome completeness in fungal genomes. A comparison of genome completeness by multiple software tools using initial and latest genome assembly versions of 45 fungi. Genome completeness expressed as a percentage of expected markers (y-axis) is plotted against assembly size in megabases (x-axis).