Projects
Software and Datasets Developed in the Lab (still being updated)
Featured
Automated genome annotation pipeline that includes self-training, RNAseq incorporation in training and prediction, functional annotation, and comparative genomics. Developed by Jon Palmer with input from Jason Stajich

SOPs for Data analyses
Automated genome assembly for simple genome projects (best for Haploids)

Phylogenomic package using BUSCO or custom protein HMMs
Genomics Tools

BioPerl project code

Automated genome annotation pipeline that includes self-training, RNAseq incorporation in training and prediction, functional annotation, and comparative genomics. Developed by Jon Palmer with input from Jason Stajich

SOPs for Data analyses

Automated genome assembly for simple genome projects (best for Haploids)
Low/Medium Coverage Zygo Genome assembly and annotation
1000 Fungal genomes project
Tools for calculating summary statistics for public fungal genomes in NCBI and downloading genomes and annotations
Protein HMMs for several public phylogenomic marker sets for Fungal Phylogenomics

Phylogenomic package using BUSCO or custom protein HMMs

An alternate tool for genome completeness that incorporates coding and non-coding elements
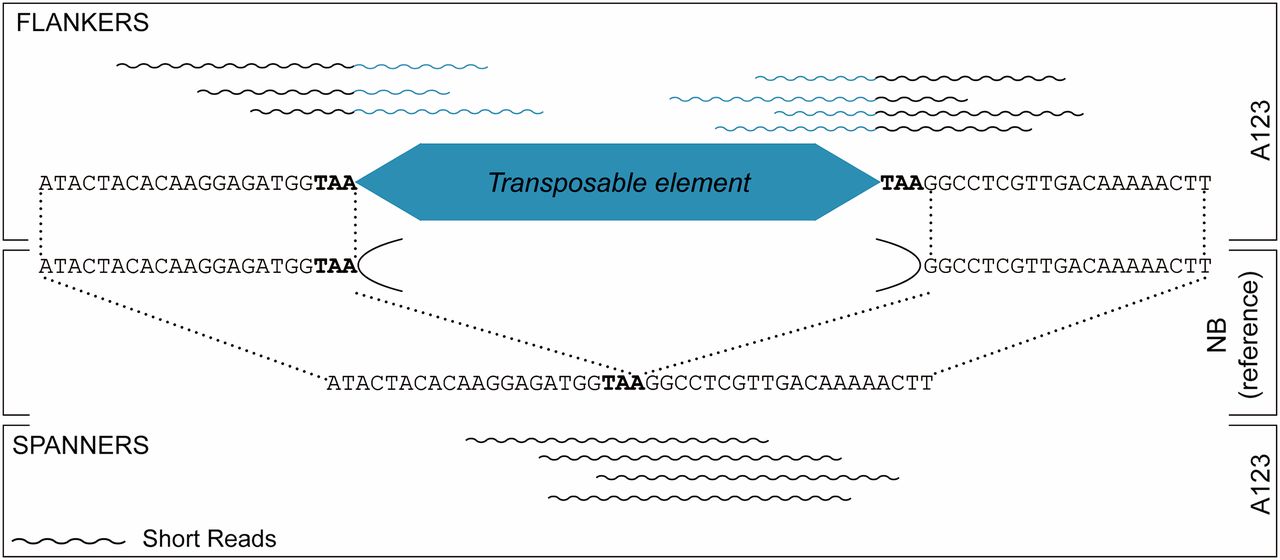
Transposable element insertion site mapping tool using short-reads for population resequencing
Datasets

BioPerl project code

Automated genome annotation pipeline that includes self-training, RNAseq incorporation in training and prediction, functional annotation, and comparative genomics. Developed by Jon Palmer with input from Jason Stajich

SOPs for Data analyses

Automated genome assembly for simple genome projects (best for Haploids)

Low/Medium Coverage Zygo Genome assembly and annotation
1000 Fungal genomes project
Tools for calculating summary statistics for public fungal genomes in NCBI and downloading genomes and annotations
Protein HMMs for several public phylogenomic marker sets for Fungal Phylogenomics

Phylogenomic package using BUSCO or custom protein HMMs

An alternate tool for genome completeness that incorporates coding and non-coding elements

Transposable element insertion site mapping tool using short-reads for population resequencing