Recent Lab Papers (2018)
There have been several manuscripts are now out from work that was done over the last few years. It has been great to be able to collaborate with many research teams as part of this work.Been slow to mention these but wanted to make sure and call out the hard work from everyone. A long term collaboration lead by Cene Gostinčar on population genomics in Hortaea, an ascomycete black yeast, complemented some reference genome work we published last year [1].
Former postdoc Ousmane Cissé published his work on Pneumocystis at the NIH in his current postdoc[2].

Current postdoc Yan Wang published part of his thesis work and we worked on these data a bit as part of the zygolife project here [3].
Graduate students Nat Pombubpa collaborated with Plant Sciences and Jeff Diez lab graduate student Courtney Collins on her thesis work looking at the mycobiome differences of soils in an alpine gradient with and without invasive shrub introduction [4].
Andrii Gryganskyi’s work on Rhizopus phylogeny with some of the first multi-genome work on this genus that was one of the earliest collaborations on some ZyGoLife questions [5].
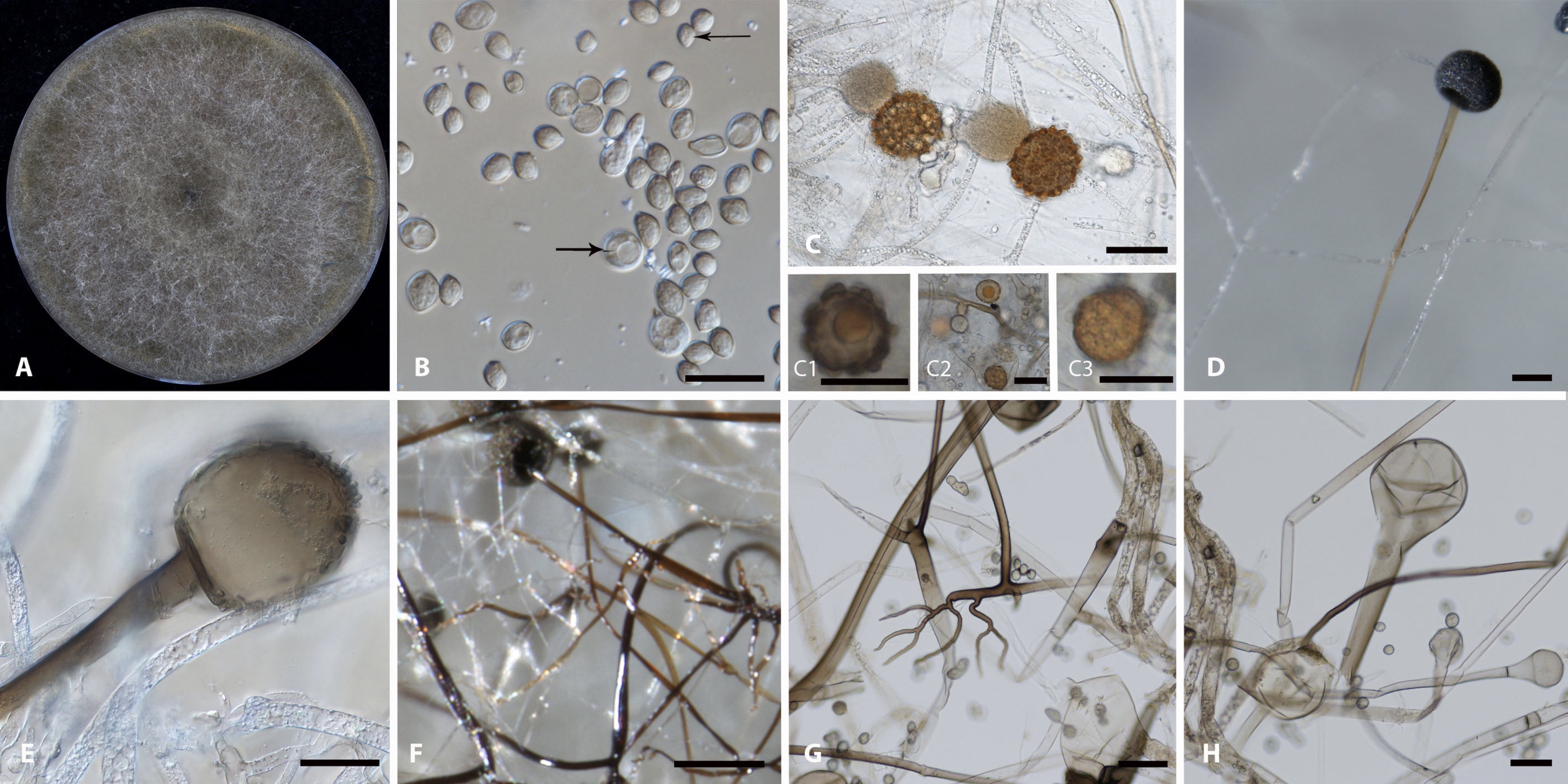
Work exploring whether Transposable elements have a role in stage-specific gene expression in Coccidioides with Theo Kirkland and Anna Muszewska [6].
A collaboration lead by Tom Richard’s lab and Guy Leonard sequence and assembled and annotated a genome of a ‘pseudofungus’ hyphochytriomycete called Hyphochytrium catenoides [7].
I think I forgot to also call out that last year postdoc Jinfeng Chen published work collaboratively with postdocs Lu Lu and Sofia Robb on the active Rice transposable element system mPing and Ping [8].
Good work all!
- Gostinčar C, Stajich JE, Zupančič J, Zalar P, Gunde-Cimerman N. Genomic evidence for intraspecific hybridization in a clonal and extremely halotolerant yeast.BMC Genomics. 2018 May 15;19(1):364. doi: 10.1186/s12864-018-4751-5.
- Wang Y, Stata M, Wang W, Stajich JE, White MM, Moncalvo JM. Comparative Genomics Reveals the Core Gene Toolbox for the Fungus-Insect Symbiosis. MBio. 2018 May 15;9(3). pii: e00636-18. doi: 10.1128/mBio.00636-18.
- Cissé OH, Ma L, Wei Huang D, Khil PP, Dekker JP, Kutty G, Bishop L, Liu Y, Deng X, Hauser PM, Pagni M, Hirsch V, Lempicki RA, Stajich JE, Cuomo CA, Kovacs JA. Comparative Population Genomics Analysis of the Mammalian Fungal Pathogen Pneumocystis. MBio. 2018 May 8;9(3). pii: e00381-18. doi: 10.1128/mBio.00381-18.
- Gryganskyi AP, Golan J, Dolatabadi S, Mondo S, Robb S, Idnurm A, Muszewska A, Steczkiewicz K, Masonjones S, Liao HL, Gajdeczka MT, Anike F, Vuek A, Anishchenko IM, Voigt K, de Hoog GS, Smith ME, Heitman J, Vilgalys R, Stajich JE. Phylogenetic and Phylogenomic Definition of Rhizopus Species. G3 (Bethesda). 2018 Apr 19. pii: g3.200235.2018. doi: 10.1534/g3.118.200235.
- Collins CG, Stajich JE, Weber SE, Pombubpa N, Diez JM. Shrub range expansion alters diversity and distribution of soil fungal communities across an alpine elevation gradient. Mol Ecol.2018 Apr 19. doi: 10.1111/mec.14694.
- Kirkland TN, Muszewska A, Stajich JE. Analysis of Transposable Elements in Coccidioides Species. J Fungi (Basel). 2018 Jan 19;4(1). pii: E13. doi: 10.3390/jof4010013.
- Leonard G, Labarre A, Milner DS, Monier A, Soanes D, Wideman JG, Maguire F, Stevens S, Sain D, Grau-Bové X, Sebé-Pedrós A, Stajich JE, Paszkiewicz K, Brown MW, Hall N, Wickstead B, Richards TA. Comparative genomic analysis of the ‘pseudofungus’ Hyphochytrium catenoides. Open Biol. 2018 Jan;8(1). pii: 170184. doi: 10.1098/rsob.170184.
- Lu L, Chen J, Robb SMC, Okumoto Y, Stajich JE, Wessler SR. Tracking the genome-wide outcomes of a transposable element burst over decades of amplification. Proc Natl Acad Sci U S A. 2017 Dec 5;114(49):E10550-E10559. doi: 10.1073/pnas.1716459114.